Topic: Computational Systems Biology – analyzing, computing and modeling biological systems
BIOSYM will become an interactive bio-modeling and simulation training course. Its modules incorporate quantitative mathematical approaches into life sciences curricula and familiarize students with the power of modeling biological processes and systems. It aims at learning how to handle and mine large data sets, analyzing and visualizing data and importing them into biocomputational modeling and simulation systems.
Computational Systems Biology with BIOSYM is a logical step towards synthesizing details and fragments of knowledge into a more holistic view of biology, and it can serve as a motivation to deal with the complexity inherent to many biological systems. BIOSYM requires an interdisciplinary approach involving biology, mathematics and computer sciences and it has a broad perspective which reaches into biomedical, pharmaceutical and biotechnological applications. It also serves basic research in fields from epidemiology to integrated human health and molecular cell biology.
BIOSYM is based on biological, chemical and physical principles applied to the conceptual teaching and learning of systems biology. In order to make the best use of the wealth of information which is and will become available about biological systems, it is essential to apply mathematical models as learning and research tools at all levels of biological education.
BIOSYM introduces students to the craft of model building, shows them how to design mathematical models and trains them to use simulations skillfully. It emphasizes the development of mathematical algorithms for the construction of simulation models and the optimization of biological processes and systems based on modeling results. The heart of BIOSYM are modules which are tailored to acquire modeling skills for specialized topics. BIOSYM will start by introducing deterministic dynamic models (e.g. growth, predator-prey interactions, selection-adaptation, enzyme kinetics etc.), advance to more complex models (e.g. epidemics, metabolic networks and control, physiology, gene regulation etc.) and introduce, on an advanced level, stochastic models and finite elements which can assist students in designing quantitative experiments with proper boundary conditions and handling large data sets (e.g. analysis of metabolomes, genome analysis, community ecology). Available data sets from hospitals (e.g. antibiotic resistances), from WHO (e.g. infectious diseases, epidemics) and from pharmaceutical firms (e.g. pharmacodynamics, mechanistic clinical models, population responses) will be used to link the modeling training to real life applications.
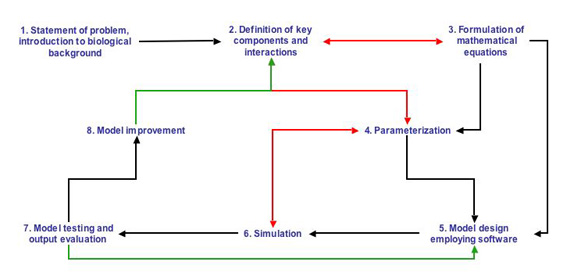
BIOSYM training is primarily based on MATLAB and its tool boxes as well as on other mathematical software (e.g. Finite Elements). Each model is introduced by a short summary of the biological facts and references to the appropriate literature, and each model contains a Java Applet or a Flash animation which will motivate the student to learn the details of the background. The actual models are then designed in MATLAB SIMULINK or another appropriate modeling language. The model architecture follows the steps as outlined in the figure
 Courses will be organized and managed via the OLAT learning platform
( http://www.olat3.unizh.ch ),
and lecture contents can be made available as BREEZE presentation documents
on the Switch BREEZE server ( http://viti.switch.ch/ )
or by using a similar software.
Courses will be organized and managed via the OLAT learning platform
( http://www.olat3.unizh.ch ),
and lecture contents can be made available as BREEZE presentation documents
on the Switch BREEZE server ( http://viti.switch.ch/ )
or by using a similar software.
BIOSYM is an interactive, blended learning package. The introduction to mathematics and the natural sciences fields, which are prerequisites for BIOSYM, can take place at any institution of higher learning according to individual curricular preferences. BIOSYM will summarize the key preconditions in an introductory module and rely on introductory mathematics and physics courses for the basics. With this background knowledge the interested student will be able to follow course programs under the guidance of a local course advisor. The advisor will recommend and supervise the modeling training scientifically and technically, organize the group meetings locally, correct exercises and tests for his/her students and improve BIOSYM by adding new ideas for novel models and developing new computational concepts.
Student who have passed the core program can flexibly choose directions which meet their goals and needs best. They may follow guided programs or develop their own computational solutions to novel problems independently and apply them to scientific research. The evaluation and credit assignments for compulsory and non-compulsory course work and its acceptance as partial fulfillments for a particular degree follows the rules which are valid at the school of inscription.
If BIOSYM proofs to be attractive to our students and if interest grows Europe-wide, we intend to offer its contents as an “International Computational Systems Biology Modeling Course” in 2008 (4 ECTS). This course would be open to selected students who have passed the basic on-line BIOSYM course. Application would be competitive and support would be needed through a special EU training course grant. The course would assemble up to 20 students for 2 weeks and contributors to BIOSYM and other qualified teachers would be invited as lecturers and to guide practical exercises.